Written by Nascent Transcript writer Brooke Wolford
November 2018
What scientific themes did you identify at the ASHG 2018 Annual Meeting? You may remember several themes, including the need for genetics research in diverse populations, the power of large cohorts like the UK Biobank, and the applications of polygenic risk scores (PRS).
A PRS, sometimes known as a genetic risk score (GRS) or genome-wide polygenic score (GPS), is a weighted sum of the effect sizes of genetic variants on a given trait as estimated from a genome wide association study (GWAS). A PRS is calculated per person, , with markers being chosen in various ways depending on the study methodology.
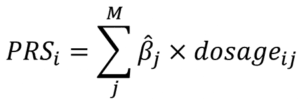
Methods like LDPred, PRSice, or metaGRS are used to calculate PRS for genotyped individuals given summary statistics from GWAS in another population. Calculation of PRS began nearly a decade ago in psychiatric traits and coronary heart disease, among others. With the availability of summary statistics from large GWAS studies, researchers are calculating and applying PRS in novel ways. Identifying the optimal PRS construction will be an important step for translation to the clinic.
In an invited session on the Clinical Implementation of Polygenic Risk Scores, 2018 Curt Stern Award winner, Sekar Kathiresan, MD, highlighted recent work demonstrating that UK Biobank participants whose genome-wide polygenic score for coronary artery disease (CAD) scored in the top 5% have greater than threefold risk for CAD compared to the rest of the population. This is similar to the CAD risk conferred by monogenic mutations, such as those causing familial hypercholesterolemia (LDLR, APOB, and PCSK9); yet 20 times as many people fall into this high-risk category as carry a monogenic mutation. He also presented unpublished work evaluating a PRS for weight in individuals with longitudinal weight measurements from the Framingham Heart Study. 20% of individuals in the top decile of the PRS distribution and 0% in the bottom decile developed severe obesity during the 26 years of follow-up.
To study how PRS could impact clinical management and outcomes, researchers in Finland have used PRS and traditional cardiovascular disease (CVD) risk factors to communicate personalized 10 year CVD risk to thousands of Finns in the GeneRISK Study, via the KardioKompassi web portal. Elisabeth Widén, MD, explained that 35% of individuals at high CVD risk took at least one action in response to their disease risk (weight loss, smoking cessation, or a doctor’s visit) as compared to just 20% of low CVD risk individuals. Furthermore, 15% of high CVD risk individuals reported smoking cessation in comparison to the 4% annual smoking cessation rate in the general population. Continued follow-up in this cohort will provide a valuable example for the introduction of PRS into clinical care and the potential public health impacts.
Lest we get too excited by the promise of PRS in precision medicine, platform talks in the Biases of PRS session urged caution in development and application of PRS. Doctoral student Sini Kerminen demonstrated that a PRS for height, generated from summary statistics from the GIANT consortium, predicted an unreasonably large difference in height between Western and Eastern Finns of 3.52 cm compared to the expected 1.6 cm, thus suggesting the accumulation of biases in PRS potentially due to uncontrolled population stratification in previous studies.
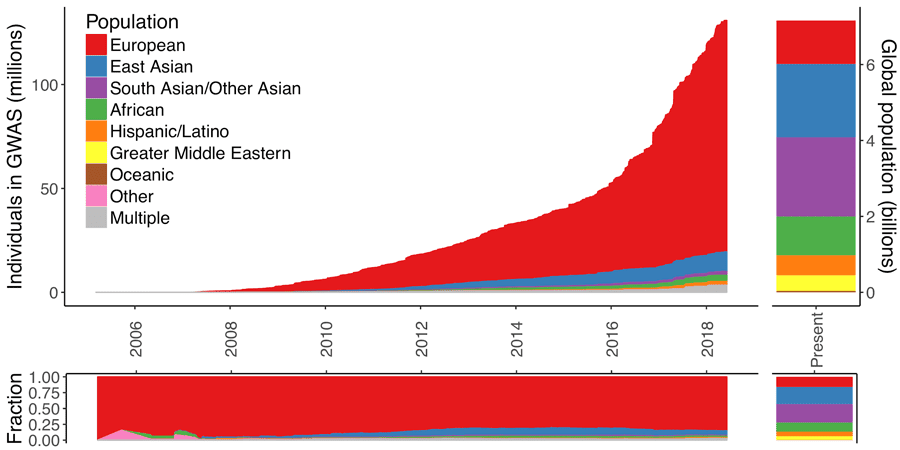
The Euro-centric GWAS bias, as highlighted in a platform talk by Alicia Martin, PhD, is a barrier for bringing the power of PRS to the clinic. As of 2016, only 19% of GWAS studies were in non-European populations, and Dr. Martin’s previous work showed these studies have limited use in diverse populations. Indeed, Bárbara Bitarello, PhD, demonstrated that based on European-ancestry GWAS summary statistics, PRS had a poorer individual level predictive power in an African American population compared to a European American population. The lack of summary statistics from GWAS in large populations of non-European ancestry means systematically biased PRS could exacerbate health disparities in already vulnerable populations.
In light of this work, it’s important to note ASHG’s position regarding diversity in genomics research cohorts:
The lack of diversity within participant cohorts in genetics and genomics research limits ability to study variation across the human genome and the genetic factors that influence health and disease, as well as our ability to ensure that every segment of the population is able to benefit from advances stemming from the research. The Society supports research policies that foster the recruitment of participants from diverse populations in genetics research. As more genetically tailored therapies come to market, it is important that the evidence base allows for their appropriate administration in all patients to avoid exacerbating health disparities.
The Euro-centric GWAS bias and other ethical concerns regarding PRS are important topics of discussion for the Society moving forward. As the predictive power of PRS continues to increase, we need to define appropriate uses of PRS: Would we predict the risk for a panel of diseases in a newborn? When would knowing one’s PRS enhance health rather than increase anxiety? My PRS take home from ASHG 2018: With great predictive power comes great responsibility.