Author: Jeffrey Calhoun, PhD
One needs only to re-watch that famous scene in the classic movie The Princess Bride to see that the concept of poison can be as fascinating as it is deadly. The term “poison” evokes a killer chemical made by human hands, but most poisons are actually naturally occurring, such as the tetrodotoxin (TTX) found in the pufferfish Fugu. Many people are willing to assume the risk of consuming Fugu just to experience the feeling of TTX even though it can be present in lethal quantities when the fish is improperly prepared.
Unlike the Fugu genome, while the human genome does not directly code for a poison per se, it does contain so-called poison exons, or small exons with in-frame premature stop codons. It is estimated that as many as one third of all genes contain at least one poison exon. These poison exons, when included in the mRNA transcript via alternative splicing, lead to nonsense mediated decay.
The discovery of poison exons, like any major discovery, produced more questions than answers. Chief among them: why would a cell waste precious energy to produce transcripts containing poison exons? Would it not be more efficient to simply reduce transcription instead? Many laboratories are hard at work trying to answer these questions, and while we don’t have a definitive answer, we have learned a tremendous amount about the role of poison exons in both normal physiology and pathophysiology.
Poison exons are smaller than average exons and are highly conserved, suggesting important functions not just in human biology. The inclusion of poison exons is involved in gene regulation, particularly during brain development. Poison exon inclusion is regulated by RNA binding proteins, and some RNA binding protein genes themselves contain poison exons as a mechanism for self-regulation. Some genes, such as FLNA, show high levels of poison exon inclusion in mature neurons relative to neuronal precursors, while others like SCN1A show the opposite pattern. In addition to its role in brain development, poison exon inclusion has been shown to be critical for a number of other biological processes including embryogenesis and spermatogenesis.
A potential role for poison exons in disease remained elusive for some time as these exons are not commonly well-annotated. Evidence suggests that genetic variants in and near poison exons can contribute to disease susceptibility. For instance, de novo variants in a poison exon lead to increased inclusion and subsequent SCN1A haploinsufficiency in multiple individuals with Dravet syndrome, a severe monogenic pediatric epilepsy primarily caused by de novo SCN1A coding variants. In addition, a recent study demonstrated roles for poison exons in cell fitness as well as tumor suppressor activity.
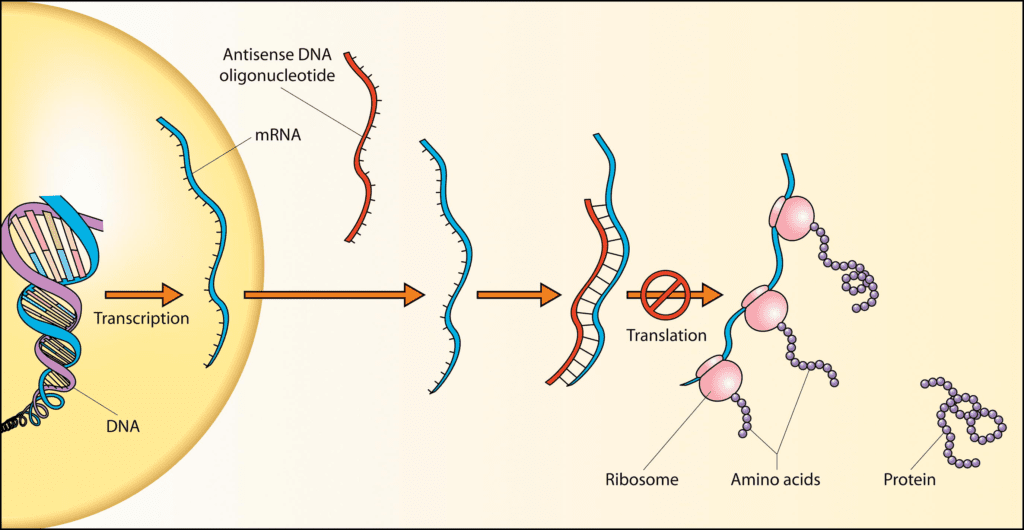
Based on their role in gene regulation, poison exons are potential targets for developing novel therapeutics. Early successes of using antisense oligonucleotides (ASOs) to modulate alternative splicing as a therapeutic include Nusinersen for spinal muscular atrophy and Milasen as a custom “N of 1” drug to correct a splicing defect in a child with Batten’s disease. While they do not target poison exons specifically, the success of these ASOs suggests they will have additional utility for targeting poison exons in other indications. In fact, a recent study using ASOs targeting poison exons by Stoke Therapeutics, Inc. showed proof-of-concept in vivo up-regulation of mRNA and protein levels by inhibition of poison exon inclusion. This technique could be applicable to Dravet syndrome or more broadly to any monogenic disorder caused by haploinsufficiency of a gene. We will hopefully have an antidote in the near future for when things go awry with this fascinating naturally occurring “poison” in our genome.
References:
Carvill GL, Mefford HC. Poison exons in neurodevelopment and disease [published online ahead of print, 2020 Jun 29]. Curr Opin Genet Dev. 2020;65:98-102. doi:10.1016/j.gde.2020.05.030
Popp MW, Maquat LE. Organizing principles of mammalian nonsense-mediated mRNA decay. Annu Rev Genet. 2013;47:139-165. doi:10.1146/annurev-genet-111212-133424
Carvill GL, Engel KL, Ramamurthy A, et al. Aberrant Inclusion of a Poison Exon Causes Dravet Syndrome and Related SCN1A-Associated Genetic Epilepsies. Am J Hum Genet. 2018;103(6):1022-1029. doi:10.1016/j.ajhg.2018.10.023
Thomas JD, Polaski JT, Feng Q, et al. RNA isoform screens uncover the essentiality and tumor-suppressor activity of ultraconserved poison exons. Nat Genet. 2020;52(1):84-94. doi:10.1038/s41588-019-0555-z
Kim J, Hu C, Moufawad El Achkar C, et al. Patient-Customized Oligonucleotide Therapy for a Rare Genetic Disease. N Engl J Med. 2019;381(17):1644-1652. doi:10.1056/NEJMoa1813279
Lim KH, Han Z, Jeon HY, et al. Antisense oligonucleotide modulation of non-productive alternative splicing upregulates gene expression. Nat Commun. 2020;11(1):3501. Published 2020 Jul 9. doi:10.1038/s41467-020-17093-9
